A quick overview of DNA and DNA sequencing - ILMN, PACB and ONT

In this article, we will focus on the basics of DNA and DNA sequencing by looking at the three different sequencing technologies on the market.
DNA, or deoxyribonucleic acid, is the instruction set for all living organisms. DNA is made up of building blocks called nucleotides. There are four of these: adenine (A), thymine (T), guanine (G), and cytosine (C). Similarly, RNA is made up of four building blocks as well, A, U (similar to T), G and C. These nucleotides are arranged in groups of three (eg. ATG, GAG etc.) and the grouping gives rise to amino acids. There are a total of 20 individual Amino Acids. The amino acids are arranged in specific patterns to give proteins. Just to understand the scale of diversity, imaging a protein is made up with 10 amino acids. The total number of possible permutations for this protein would thus be 1020. This large number of permutations thus allows individual proteins to be unique.
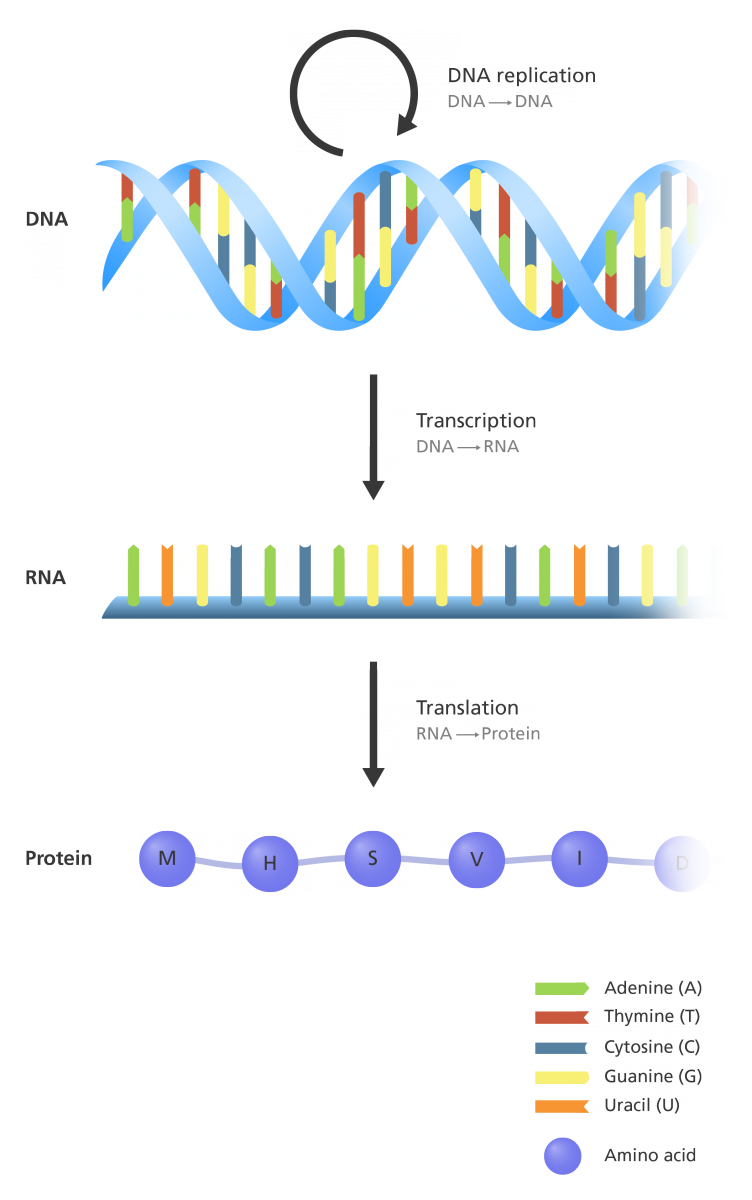
The sequence of life
So, what does sequencing entail?
Sequencing is the process of determining the DNA sequence of an organism. This is important because DNA determines the RNA sequence which determines what proteins are made, and proteins are the workhorses of cells.
The genome refers to the complete DNA set of an organism. For example, the human genome would entail all the DNA sequences found in you or me. If you’re about to punch your computer, fret not, I am done with all the technical details – for now.
Sequencing Human's - The Human Genome Project
Back in 2000, we set about an ambitious plan to sequence the human genome through the "The Human Genome Project". It was estimated that it would cost US$3 billion and 15 years to sequence the whole human genome. But, the project finally finished in 2022 when the first gap-free full human genome sequence was published. A full new article covering this can be found here: https://www.theguardian.com/science/2022/mar/31/first-complete-gap-free-human-genome-sequence-published
Over the last 22 years, as DNA sequencing has become common place in many laboratories and medical facilities, the cost of sequencing a human genome has fallen to under US$1000. What’s more, the sequencing can be done in days or even hours.
Let’s have a look at three companies in this field – Illumina (NASDAQ: ILMN), Pacific Biosciences (NASDAQ: PACB), and Oxford Nanopore (LON: ONT) to understand how their technologies help in determining the DNA sequence.
Analyzing Illumina’s technology - the leader in short read sequencing
Let’s start with Illumina. The company’s machines allow researchers and clinicians to sequence ‘short fragments’ of an individual’s genome. Illumina’s technology is based on fluorescence technology, where each nucleotide is encoded by a specific colour. To sequence a genome, the genome is first cut into multiple fragments. Each fragment is then copied multiple times to create a cluster. Once a cluster is produced, the sequencing starts. Each cluster is then monitored at the same time by an optical system based on the fluorescence which is emitted, which allows a sequence to be determined.
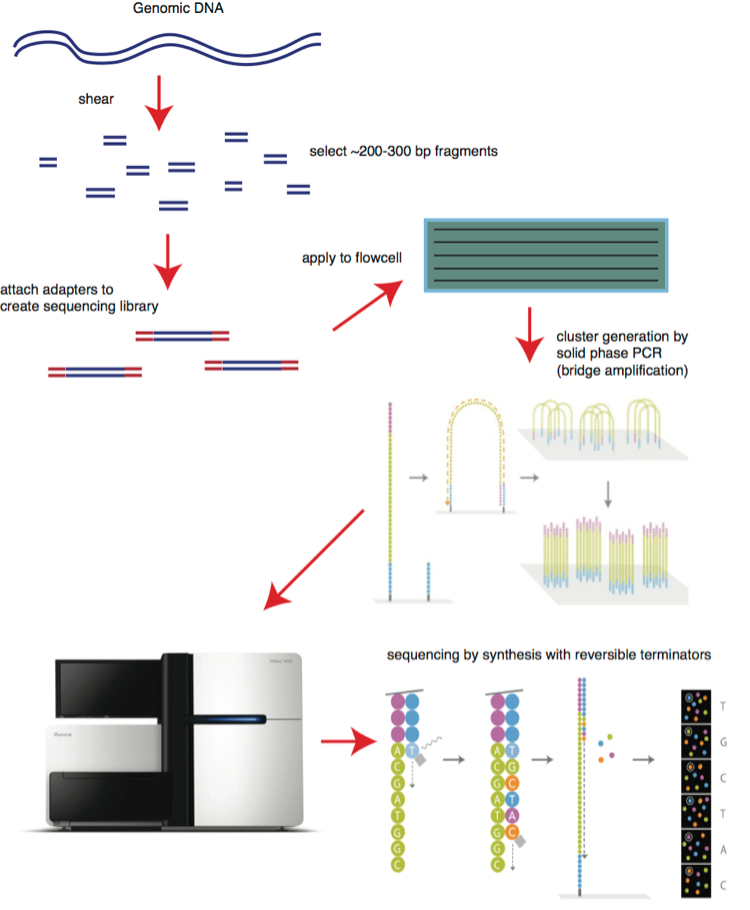
The brief explanation of how an Illumina machine works bring up two questions:
1) How are the small fragments put back together?
This is one of the advantages and limitations of the system. The advantage is that by having short fragments, researchers and clinicians can shorten the time required for sequencing by focusing only on segments that they are interested in. This means that the whole genome need not be sequenced all the time – instead, only proteome sequencing might be required. Proteome is the complete set of proteins encoded by a particular genome.
The limitation of having to work with small fragments appears when a long DNA strand or whole genome needs to be sequenced. After sequencing is performed, the data needs to be put together, which eats up computing power. This means that it can take another few days before the final sequencing results can be used. The need to piece together the short fragments also causes another issue: The need for a reference sequence which the computer uses to align all the sequences together.
2) Why do clusters need to be formed?
Clusters are an important methodology which Illumina developed. Clusters allow Illumina’s machines to ensure that the sequencing data generated is reliable and accurate. This is because if 100 of the same fragments are sequenced at the same time, the probability of a mistake is reduced. This technology gives Illumina’s machines high reliability and accuracy, and these traits have helped the company gain market share.
Understanding Pacific Bioscience’s technology
Pacific Biosciences works with fluorescence technology, just like Illumina. However, in Pac Bio’s system, the DNA strands are not cut into fragments. Rather Pac Bio’s system allows researchers to do what is known as “long fragment” sequencing. So, what are the advantages and limitations of this system?
One of the biggest advantages of long reads is that it allows sequencing of new organisms – that is much harder to do on Illumina’s system. By having long reads, Pac Bio’s system removes the need to piece together the fragments after sequencing. Pac Bio’s system is preferred over Illumina’s when new organisms are being sequenced as a reference sequence is not available.
A key problem faced by the Pacific Biosciences platform is that the platform doesn't perform parallel sequencing like Illumina. In its initial years this hindered the reliability and accuracy of the platform, because of the weak signal strength generated by a single fluorophore during base incorporation. However, this issue was addressed by the company in the last few years when it developed the SMRT technology that allowed it to sequence a single strand multiple times. The solution was an elegant one which saw the accuracy of Pacific Biosciences sequencing readout improve by multiple folds resulting in 99% accuracy. The one downside thou is the slightly lower output generated by the platform.
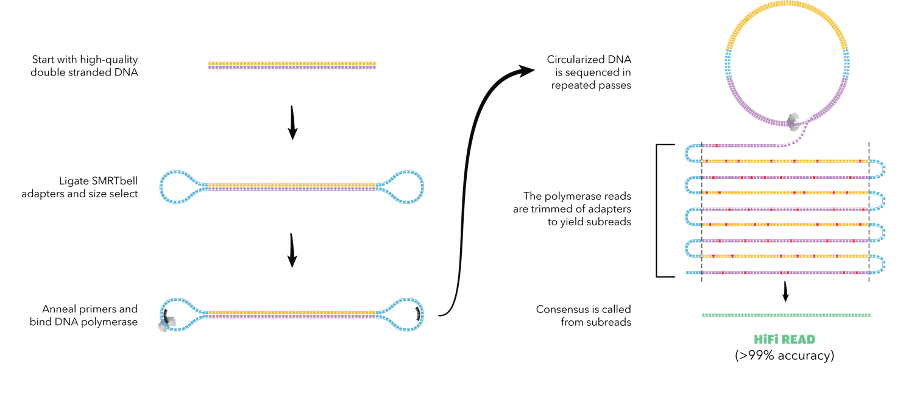
Poring over Oxford Nanopore’s technology
Oxford Nanopore’s system works differently, as it’s based on disruptions in electrical impulses for sequencing. As a DNA strand moves through a nanopore (i.e. a nano-sized hole), it causes a disruption in the set electrical pulse of the nanopore. This disruption makes it possible to identify the nucleotide through decoding by an algorithm.
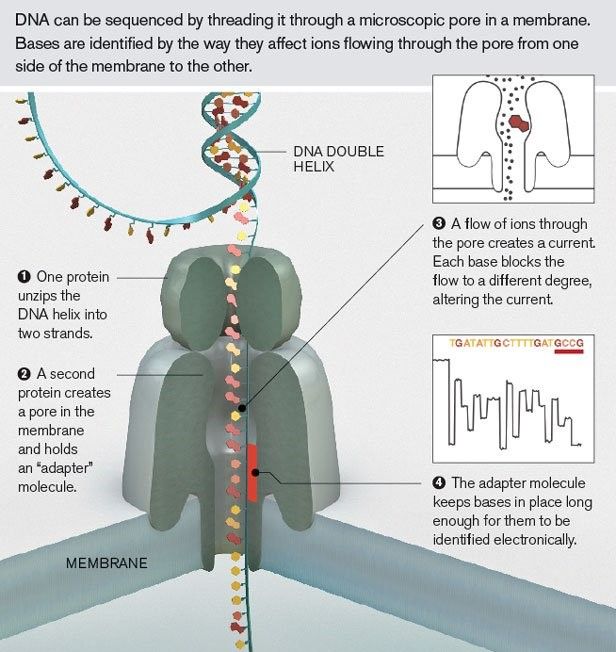
While a few years back, the accuracy of Oxford Nanopore was a huge bottleneck. This seems to be less of an issue now. In 2020, Oxford Nanopore revealed a number of key advances for its sequencing technology. It shared that a new analysis algorithm and chemistry for its system improved the single read accuracy to 99.1% while structural variant accuracy was at 96% (Gold standard) and SNP accuracy stood at 99.92% (similar to short read). It also shared that they successfully sequenced a record 10 Tb in a single run. The super long reads allowed by the nanopore system is clearly an advantage that the nanopore system has over its competitors. (Source: At NCM, announcements include single-read accuracy of 99.1% on new chemistry and sequencing a record 10 Tb in a single PromethION run (nanoporetech.com)
Additionally, just a few day ago (30 Mar 2022) Oxford Nanopore announced that its accuracy further improved to 99.3%. It also announced new features for the Nanopore which allows real time methylation analysis and a new Short Fragment Mode which allows users to sequence fragments as short as 20 bases.
Oxford Nanopore’s technology advantages go further: The tiny size of the company’s sequencing system allows researchers who are not working in a lab to sequencing on the go. For example, imagine a marine biologist who is out at sea and needs to sequence a rare fish he just caught. The Oxford Nanopore system will allow him to perform sequencing quickly even if he’s out at sea – the convenience of Oxford Nanopore’s product is a huge advantage as it allows researchers to make decisions quickly.
Let’s wrap it up here for this article and stay tuned for the next article in which we will explore the applications of DNA sequencing.
If you liked our article, subscribe to our newsletter to receive our latest articles directly in your inbox. The subscribe button can be found at the corner of the page. We will appreciate your support, or hit the follow button on twitter or Linkedin.
Disclaimer: All opinions shared in this article are the opinions of the authors and do not constitute financial advice or recommendations to buy or sell. Please consult a financial advisor before you make any financial decisions.





Comments ()